Notebook source code:
examples/bacteria/bacteria.ipynb
Run the notebook yourself on binder

Optimal resource allocation in microbial growth¶
Sacha PSALMON, Baptiste SCHALL, Enzo ISNARD and Guillaume GROS
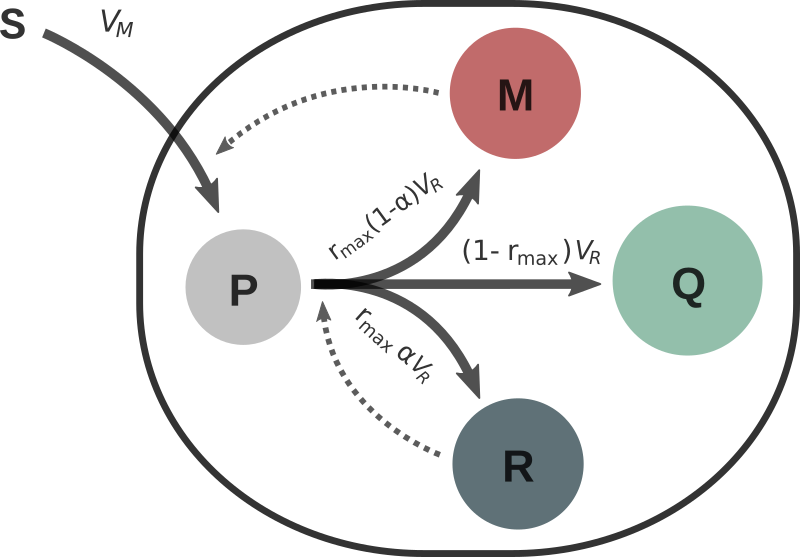
Microorganisms must assign the resources available in the environment to different cellular functions. In nature, it is assumed that bacteria have evolved in a way that their resource allocation strategies maximize their biomass. Consider the simple resource allocation mathematical model
where \(p\), \(r\), \(m\) and \(q\) are the mass fractions of: precursor metabolites, the gene expression machinery (ribosomes, RNA polymerase…), the metabolic machinery (enzymes, transporters…) and the housekeeping machinery respectively. The microbial growth rate is defined as \(v_R(p,r) = \frac{p(r-r_{min})}{K+p}\). Constants \(E_M\), \(r_{max}\), \(r_{min}\) and \(K\) are positive. Maximizing the bacterial biomass is equivalent to maximizing the integral of the growth rate over a fixed time window \([0,T]\). Thus, the cost function of the optimal control problem can be written as
with the resource allocation parameter (i.e. the control) constrained to \(\alpha(t) \in [0,1]\).
[1]:
import matplotlib.pyplot as plt
from matplotlib.animation import FuncAnimation
import matplotlib.animation as animation
from IPython.display import HTML
%matplotlib inline
import numpy as np
import math
import os,shutil
!pygmentize ./problem.cpp
// +++DRAFT+++ This class implements the OCP functions
// It derives from the generic class bocop3OCPBase
// OCP functions are defined with templates since they will be called
// from both the NLP solver (double arguments) and AD tool (ad_double arguments)
//#pragma once
#include <OCP.h>
// ///////////////////////////////////////////////////////////////////
template <typename Variable>
void OCP::finalCost(double initial_time, double final_time, const Variable *initial_state, const Variable *final_state, const Variable *parameters, const double *constants, Variable &final_cost)
{
// maximize the volume of the bacteria
final_cost = -final_state[3];
}
template <typename Variable>
void OCP::dynamics(double time, const Variable *state, const Variable *control, const Variable *parameters, const double *constants, Variable *state_dynamics)
{
Variable p = state[0];
Variable r = state[1];
Variable m = state[2];
Variable V = state[3];
Variable u = control[0];
double Em = constants[0];
double K = constants[1];
double rmax = constants[2];
double rmin = constants[3];
Variable vR = (r-rmin)*p/(K+p);
state_dynamics[0] = Em*m - (p+1)*vR;
state_dynamics[1] = (u*rmax-r)*vR;
state_dynamics[2] = ((1-u)*rmax-m)*vR;
state_dynamics[3] = vR*V;
}
template <typename Variable>
void OCP::boundaryConditions(double initial_time, double final_time, const Variable *initial_state, const Variable *final_state, const Variable *parameters, const double *constants, Variable *boundary_conditions)
{
boundary_conditions[0] = initial_state[0];
boundary_conditions[1] = initial_state[1];
boundary_conditions[2] = initial_state[2];
boundary_conditions[3] = initial_state[3];
}
template <typename Variable>
void OCP::pathConstraints(double time, const Variable *state, const Variable *control, const Variable *parameters, const double *constants, Variable *path_constraints)
{}
void OCP::preProcessing()
{}
// ///////////////////////////////////////////////////////////////////
// explicit template instanciation for template functions, with double and double_ad
// +++ could be in an included separate file ?
// but needs to be done for aux functions too ? APPARENTLY NOT !
template void OCP::finalCost<double>(double initial_time, double final_time, const double *initial_state, const double *final_state, const double *parameters, const double *constants, double &final_cost);
template void OCP::dynamics<double>(double time, const double *state, const double *control, const double *parameters, const double *constants, double *state_dynamics);
template void OCP::boundaryConditions<double>(double initial_time, double final_time, const double *initial_state, const double *final_state, const double *parameters, const double *constants, double *boundary_conditions);
template void OCP::pathConstraints<double>(double time, const double *state, const double *control, const double *parameters, const double *constants, double *path_constraints);
template void OCP::finalCost<double_ad>(double initial_time, double final_time, const double_ad *initial_state, const double_ad *final_state, const double_ad *parameters, const double *constants, double_ad &final_cost);
template void OCP::dynamics<double_ad>(double time, const double_ad *state, const double_ad *control, const double_ad *parameters, const double *constants, double_ad *state_dynamics);
template void OCP::boundaryConditions<double_ad>(double initial_time, double final_time, const double_ad *initial_state, const double_ad *final_state, const double_ad *parameters, const double *constants, double_ad *boundary_conditions);
template void OCP::pathConstraints<double_ad>(double time, const double_ad *state, const double_ad *control, const double_ad *parameters, const double *constants, double_ad *path_constraints);
[2]:
import bocop
problem1_path = "." # using local problem definition
bocop.build(problem1_path, cmake_options = '-DCMAKE_CXX_COMPILER=g++')
[EXEC] > ['cmake -DCMAKE_BUILD_TYPE=Debug -DPROBLEM_DIR=/home/caillau/gallery/examples/bacteria -DCPP_FILE=problem.cpp -DCMAKE_CXX_COMPILER=g++ /home/caillau/.conda/envs/ct-gallery/lib/python3.7/site-packages/bocop']
> -- The C compiler identification is GNU 7.5.0
> -- The CXX compiler identification is GNU 9.3.0
> -- Detecting C compiler ABI info
> -- Detecting C compiler ABI info - done
> -- Check for working C compiler: /home/caillau/.conda/envs/ct-gallery/bin/x86_64-conda-linux-gnu-cc - skipped
> -- Detecting C compile features
> -- Detecting C compile features - done
> -- Detecting CXX compiler ABI info
> -- Detecting CXX compiler ABI info - done
> -- Check for working CXX compiler: /usr/bin/g++ - skipped
> -- Detecting CXX compile features
> -- Detecting CXX compile features - done
> -- Problem path: /home/caillau/gallery/examples/bacteria
> -- Using CPPAD found at /home/caillau/.conda/envs/ct-gallery/include/cppad/..
> -- Using IPOPT found at /home/caillau/.conda/envs/ct-gallery/lib/libipopt.so
> -- Found Python3: /home/caillau/.conda/envs/ct-gallery/bin/python3.7 (found version "3.7.10") found components: Interpreter Development Development.Module Development.Embed
> -- Found SWIG: /home/caillau/.conda/envs/ct-gallery/bin/swig (found suitable version "4.0.2", minimum required is "4")
> -- Build type: Debug
> -- Configuring done
> -- Generating done
> -- Build files have been written to: /home/caillau/gallery/examples/bacteria/build
[DONE] > ['cmake -DCMAKE_BUILD_TYPE=Debug -DPROBLEM_DIR=/home/caillau/gallery/examples/bacteria -DCPP_FILE=problem.cpp -DCMAKE_CXX_COMPILER=g++ /home/caillau/.conda/envs/ct-gallery/lib/python3.7/site-packages/bocop']
[EXEC] > make
> make: Warning: File 'Makefile' has modification time 10 s in the future
> make[1]: Warning: File 'CMakeFiles/Makefile2' has modification time 10 s in the future
> make[2]: Warning: File 'src/CMakeFiles/bocop.dir/flags.make' has modification time 10 s in the future
> make[2]: warning: Clock skew detected. Your build may be incomplete.
> make[2]: Warning: File 'src/CMakeFiles/bocop.dir/flags.make' has modification time 10 s in the future
> [ 3%] Building CXX object src/CMakeFiles/bocop.dir/AD/dOCPCppAD.cpp.o
> [ 7%] Building CXX object src/CMakeFiles/bocop.dir/DOCP/dOCP.cpp.o
> [ 10%] Building CXX object src/CMakeFiles/bocop.dir/DOCP/dODE.cpp.o
> [ 14%] Building CXX object src/CMakeFiles/bocop.dir/DOCP/dControl.cpp.o
> [ 17%] Building CXX object src/CMakeFiles/bocop.dir/DOCP/dState.cpp.o
> [ 21%] Building CXX object src/CMakeFiles/bocop.dir/DOCP/solution.cpp.o
> [ 25%] Building CXX object src/CMakeFiles/bocop.dir/NLP/NLPSolverIpopt.cpp.o
> [ 28%] Building CXX object src/CMakeFiles/bocop.dir/OCP/OCP.cpp.o
> [ 32%] Building CXX object src/CMakeFiles/bocop.dir/tools/tools.cpp.o
> [ 35%] Building CXX object src/CMakeFiles/bocop.dir/tools/tools_interpolation.cpp.o
> [ 39%] Building CXX object src/CMakeFiles/bocop.dir/home/caillau/gallery/examples/bacteria/problem.cpp.o
> [ 42%] Linking CXX shared library /home/caillau/gallery/examples/bacteria/libbocop.so
> make[2]: warning: Clock skew detected. Your build may be incomplete.
> [ 42%] Built target bocop
> Scanning dependencies of target bocopwrapper_swig_compilation
> [ 46%] Swig compile /home/caillau/.conda/envs/ct-gallery/lib/python3.7/site-packages/bocop/src/bocopwrapper.i for python
> Language subdirectory: python
> Search paths:
> ./
> /home/caillau/.conda/envs/ct-gallery/include/python3.7m/
> AD/
> DOCP/
> NLP/
> OCP/
> tools/
> /home/caillau/.conda/envs/ct-gallery/include/cppad/../
> /home/caillau/.conda/envs/ct-gallery/include/coin/
> /home/caillau/.conda/envs/ct-gallery/include/python3.7m/
> /home/caillau/.conda/envs/ct-gallery/lib/python3.7/site-packages/bocop/src/AD/
> /home/caillau/.conda/envs/ct-gallery/lib/python3.7/site-packages/bocop/src/DOCP/
> /home/caillau/.conda/envs/ct-gallery/lib/python3.7/site-packages/bocop/src/NLP/
> /home/caillau/.conda/envs/ct-gallery/lib/python3.7/site-packages/bocop/src/OCP/
> /home/caillau/.conda/envs/ct-gallery/lib/python3.7/site-packages/bocop/src/tools/
> ./swig_lib/python/
> /home/caillau/.conda/envs/ct-gallery/share/swig/4.0.2/python/
> ./swig_lib/
> /home/caillau/.conda/envs/ct-gallery/share/swig/4.0.2/
> Preprocessing...
> Starting language-specific parse...
> Processing types...
> C++ analysis...
> Processing nested classes...
> Generating wrappers...
> [ 46%] Built target bocopwrapper_swig_compilation
> make[2]: Warning: File 'src/CMakeFiles/bocopwrapper.dir/bocopwrapperPYTHON_wrap.cxx' has modification time 10 s in the future
> [ 50%] Building CXX object src/CMakeFiles/bocopwrapper.dir/CMakeFiles/bocopwrapper.dir/bocopwrapperPYTHON_wrap.cxx.o
> [ 53%] Linking CXX shared module /home/caillau/gallery/examples/bacteria/_bocopwrapper.so
> -- Moving python modules to /home/caillau/gallery/examples/bacteria
> make[2]: warning: Clock skew detected. Your build may be incomplete.
> [ 53%] Built target bocopwrapper
> [ 57%] Building CXX object src/CMakeFiles/bocopApp.dir/AD/dOCPCppAD.cpp.o
> [ 60%] Building CXX object src/CMakeFiles/bocopApp.dir/DOCP/dOCP.cpp.o
> [ 64%] Building CXX object src/CMakeFiles/bocopApp.dir/DOCP/dODE.cpp.o
> [ 67%] Building CXX object src/CMakeFiles/bocopApp.dir/DOCP/dControl.cpp.o
> [ 71%] Building CXX object src/CMakeFiles/bocopApp.dir/DOCP/dState.cpp.o
> [ 75%] Building CXX object src/CMakeFiles/bocopApp.dir/DOCP/solution.cpp.o
> [ 78%] Building CXX object src/CMakeFiles/bocopApp.dir/NLP/NLPSolverIpopt.cpp.o
> [ 82%] Building CXX object src/CMakeFiles/bocopApp.dir/OCP/OCP.cpp.o
> [ 85%] Building CXX object src/CMakeFiles/bocopApp.dir/tools/tools.cpp.o
> [ 89%] Building CXX object src/CMakeFiles/bocopApp.dir/tools/tools_interpolation.cpp.o
> [ 92%] Building CXX object src/CMakeFiles/bocopApp.dir/home/caillau/gallery/examples/bacteria/problem.cpp.o
> [ 96%] Building CXX object src/CMakeFiles/bocopApp.dir/main.cpp.o
> [100%] Linking CXX executable /home/caillau/gallery/examples/bacteria/bocopApp
> [100%] Built target bocopApp
> make[1]: warning: Clock skew detected. Your build may be incomplete.
> make: warning: Clock skew detected. Your build may be incomplete.
[DONE] > make
Done
[2]:
0
[3]:
path_def = './def'
fileList = os.listdir(path_def)
for file in os.listdir(path_def):
if file.endswith('.def'):
shutil.copyfile(os.path.join(path_def,file), os.path.join(problem1_path,'problem.def'))
sol_name = file.replace('.def','.sol')
bocop.run(problem1_path, graph=0)
os.rename("problem.sol",sol_name)
Executing bocop ...
Done
Executing bocop ...
Done
Executing bocop ...
Done
Executing bocop ...
Done
Optimal resource allocation¶
[4]:
rmin = 0.07
rmax = 0.5
umin = rmin/rmax
Em = 0.6; K = 0.003
param = [Em,K,rmax,rmin]
popt = np.sqrt(K*Em)
f = popt/(K + popt)*(popt+1)
uopt = (Em + f*rmin/rmax)/(Em + f)
ropt = uopt*rmax
mopt = (1-uopt)*rmax
muopt = popt/(K + popt)*(ropt-rmin)
solution = bocop.readSolution("./problem1.sol")
t = solution.time_steps
p = solution.state[0]
r = solution.state[1]
m = solution.state[2]
V = solution.state[3]
u = solution.control[0]
q = 1 - r - m
mu = p*(r-rmin)/(K + p)
t0 = t[0]; t1 = t[-1]
plt.figure(0, figsize=(10,5))
plt.subplot(221)
plt.subplots_adjust(hspace=0.3, wspace=0.5)
plt.grid(linestyle='dotted')
ax = plt.gca()
plt.xlabel('t')
plt.ylabel('Allocation $\\alpha$')
plt.plot([t0,t1],[0,0],linewidth=1,color='grey',linestyle='--')
plt.plot([t0,t1],[1,1],linewidth=1,color='grey',linestyle='--')
plt.plot([t0,t1],[uopt,uopt],linewidth=1, linestyle='dotted', color='black',label='$\\alpha^*_{opt}$')
plt.plot(t[1:],u,linewidth=1,label='$\\alpha$')
ax.set_xlim(t0,t1)
plt.tick_params(axis='x', which='both', bottom=False, top=False, labelbottom=False)
ax2 = ax.twinx()
ax2.set_ylim(ax.get_ylim())
plt.tick_params(axis='y', which='minor', labelleft='off', labelright='on')
plt.yticks([uopt], ['$\\alpha^*_{opt}$'])
plt.subplot(222)
plt.grid(linestyle='dotted')
ax = plt.gca()
plt.plot([t0,t1],[popt,popt],linewidth=1, linestyle='dotted', color='black',label='$\hat p^*_{opt}$')
plt.plot(t, p, linewidth=1, label='$p$')
plt.legend()
ax.set_xlim(t0,t1)
plt.xlabel('t')
plt.ylabel('$p$')
ax2 = ax.twinx()
ax2.set_ylim(ax.get_ylim())
plt.tick_params(axis='y', which='minor', labelleft='off', labelright='on')
plt.yticks([popt], ['$p^*_{opt}$'])
plt.subplot(223)
plt.grid(linestyle='dotted')
plt.xlabel('t')
plt.ylabel('Mass fractions')
ax = plt.gca()
ax2 = ax.twinx()
ax2.set_ylim(ax.get_ylim())
plt.tick_params(axis='y', which='minor', labelleft='off', labelright='on')
plt.yticks([rmin,ropt,rmax], ['$r_{min}$','$r^*_{opt}$','$r_{max}$'])
ax.set_xlim(t[0],t[-1])
plt.fill_between(t, r, color='#374e55', label='$r$', alpha=0.7)
plt.fill_between(t, r, r+m, color='#b24746', label='$m$', alpha=0.8)
plt.fill_between(t, r+m, 1, color='#79af97', label='$q$', alpha=0.9)
plt.plot(t, r, color='#374e55',linewidth=0.5)
plt.plot(t, r+m, color='#b24746',linewidth=0.5)
plt.grid(linestyle='dotted',color='black')
plt.legend(ncol = 2)
plt.subplot(224)
plt.grid(linestyle='dotted')
plt.xlabel('t')
plt.ylabel('Growth rate $\mu$')
ax = plt.gca()
plt.plot([t0,t1],[muopt,muopt],linewidth=1, linestyle='dotted', color='black',label='$\mu^*_{opt}$')
plt.plot(t,mu,linewidth=1,label='$\mu(p,r)$')
ax.set_xlim(t0,t1)
ax2 = ax.twinx()
ax2.set_ylim(ax.get_ylim())
plt.tick_params(axis='y', which='minor', labelleft='off', labelright='on')
plt.yticks([muopt], ['$\mu^*_{opt}$'])
plt.show()
Loading solution: ./problem1.sol
Comparison between the optimal and naive method¶
(Naive method: \(\alpha(t)\) constant for the interval \([0,T]\))
[5]:
naivesol = bocop.readSolution("./naiveproblem.sol")
length=len(solution.state[1])
langs=['r','m','q']
colors = ["#374e55","#b24746", "#79af97"]
fig = plt.figure(figsize=(12,6))
ax1 = plt.subplot(1,2,1)
ax2 = plt.subplot(1,2,2)
def update(frame):
ax1.clear()
ax2.clear()
r=solution.state[1][frame]
m=solution.state[2][frame]
q=1-r-m
rn=naivesol.state[1][frame]
mn=naivesol.state[2][frame]
qn=1-rn-mn
ax1.pie([r,m,q], colors=colors, labels=langs, autopct='%1.2f%%',startangle=90,radius=math.sqrt(solution.state[3][frame])*3+0.7)
title1='(Optimal Method) Volume = '+str(solution.state[3][frame])
ax1.set_title(title1)
ax2.pie([rn,mn,qn], colors=colors, labels=langs, autopct='%1.2f%%',startangle=90,radius=math.sqrt(naivesol.state[3][frame])*3+0.7)
title2='(Naive Method) Volume = '+str(naivesol.state[3][frame])
ax2.set_title(title2)
ani = FuncAnimation(fig, update, frames=range(length),interval=10, repeat=False)
FFwriter= animation.FFMpegWriter(fps=120)
ani.save('./images/comparison.mp4',writer=FFwriter)
HTML(ani.to_html5_video())
Loading solution: ./naiveproblem.sol
[5]:
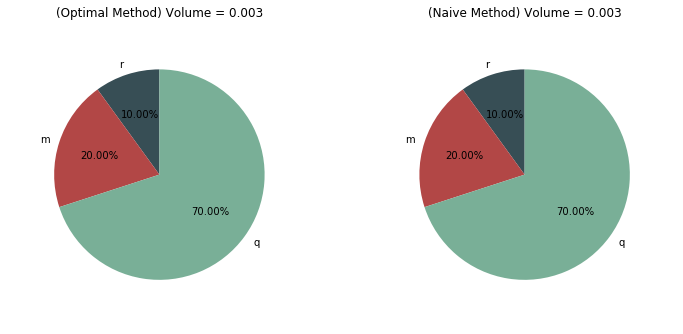
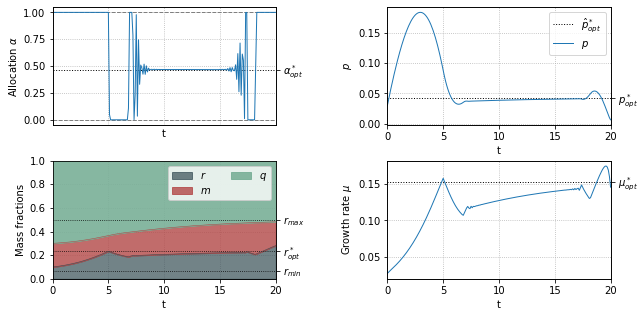
Optimal resource allocation during a nutrient shift¶
[6]:
rmin = 0.07
rmax = 0.5
umin = rmin/rmax
Em = 0.7; K = 0.003
param = [Em,K,rmax,rmin]
popt = np.sqrt(K*Em)
f = popt/(K + popt)*(popt+1)
uopt = (Em + f*rmin/rmax)/(Em + f)
ropt = uopt*rmax
mopt = (1-uopt)*rmax
muopt = popt/(K + popt)*(ropt-rmin)
solution = bocop.readSolution("./problem2.sol")
t = solution.time_steps
p = solution.state[0]
r = solution.state[1]
m = solution.state[2]
V = solution.state[3]
u = solution.control[0]
q = 1 - r - m
mu = p*(r-rmin)/(K + p)
t0 = t[0]; t1 = t[-1]
plt.figure(0, figsize=(10,5))
plt.subplot(221)
plt.subplots_adjust(hspace=0.3, wspace=0.5)
plt.grid(linestyle='dotted')
ax = plt.gca()
plt.xlabel('t')
plt.ylabel('Allocation $\\alpha$')
plt.plot([t0,t1],[0,0],linewidth=1,color='grey',linestyle='--')
plt.plot([t0,t1],[1,1],linewidth=1,color='grey',linestyle='--')
plt.plot([t0,t1],[uopt,uopt],linewidth=1, linestyle='dotted', color='black',label='$\\alpha^*_{opt}$')
plt.plot(t[1:],u,linewidth=1,label='$\\alpha$')
ax.set_xlim(t0,t1)
ax2 = ax.twinx()
ax2.set_ylim(ax.get_ylim())
plt.tick_params(axis='y', which='minor', labelleft='off', labelright='on')
plt.yticks([uopt], ['$\\alpha^*_{opt}$'])
plt.subplot(222)
plt.grid(linestyle='dotted')
ax = plt.gca()
plt.plot([t0,t1],[popt,popt],linewidth=1, linestyle='dotted', color='black',label='$\hat p^*_{opt}$')
plt.plot(t, p, linewidth=1, label='$p$')
plt.legend()
ax.set_xlim(t0,t1)
plt.xlabel('t')
plt.ylabel('$p$')
ax2 = ax.twinx()
ax2.set_ylim(ax.get_ylim())
plt.tick_params(axis='y', which='minor', labelleft='off', labelright='on')
plt.yticks([popt], ['$p^*_{opt}$'])
plt.subplot(223)
plt.grid(linestyle='dotted')
plt.xlabel('t')
plt.ylabel('Mass fractions')
ax = plt.gca()
ax2 = ax.twinx()
ax2.set_ylim(ax.get_ylim())
plt.tick_params(axis='y', which='minor', labelleft='off', labelright='on')
plt.yticks([rmin,ropt,rmax], ['$r_{min}$','$r^*_{opt}$','$r_{max}$'])
ax.set_xlim(t[0],t[-1])
plt.fill_between(t, r, color='#374e55', label='$r$', alpha=0.7)
plt.fill_between(t, r, r+m, color='#b24746', label='$m$', alpha=0.8)
plt.fill_between(t, r+m, 1, color='#79af97', label='$q$', alpha=0.9)
plt.plot(t, r, color='#374e55',linewidth=0.5)
plt.plot(t, r+m, color='#b24746',linewidth=0.5)
plt.grid(linestyle='dotted',color='black')
plt.legend(ncol = 2)
plt.subplot(224)
plt.grid(linestyle='dotted')
plt.xlabel('t')
plt.ylabel('Growth rate $\mu$')
ax = plt.gca()
plt.plot([t0,t1],[muopt,muopt],linewidth=1, linestyle='dotted', color='black',label='$\mu^*_{opt}$')
plt.plot(t,mu,linewidth=1,label='$\mu(p,r)$')
ax.set_xlim(t0,t1)
ax2 = ax.twinx()
ax2.set_ylim(ax.get_ylim())
plt.tick_params(axis='y', which='minor', labelleft='off', labelright='on')
plt.yticks([muopt], ['$\mu^*_{opt}$'])
plt.show()
Loading solution: ./problem2.sol
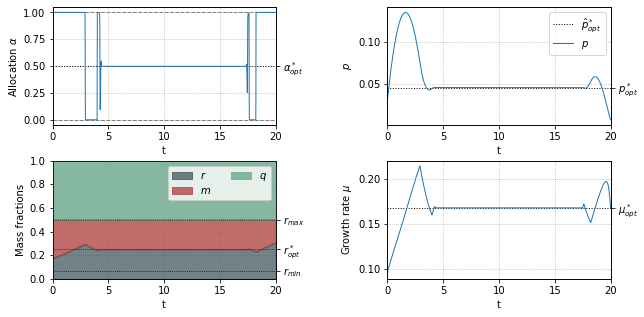
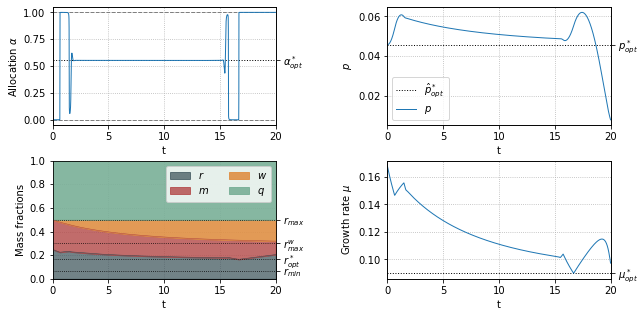
Optimal resource allocation when synthesizing an anti-stress protein \(w\)¶
[7]:
rmax = 0.3
rmax_old = 0.5
rmin = 0.07
umin = rmin/rmax
Em = 0.7; K = 0.003
param = [Em,K,rmax,rmin]
popt = np.sqrt(K*Em)
f = popt/(K + popt)*(popt+1)
uopt = (Em + f*rmin/rmax)/(Em + f)
ropt = uopt*rmax
mopt = (1-uopt)*rmax
muopt = popt/(K + popt)*(ropt-rmin)
solution = bocop.readSolution("./problem3.sol")
t = solution.time_steps
p = solution.state[0]
r = solution.state[1]
m = solution.state[2]
V = solution.state[3]
u = solution.control[0]
q = 1 - r - m
mu = p*(r-rmin)/(K + p)
t0 = t[0]; t1 = t[-1]
names = ['p','r','m','\\alpha']
plt.figure(0, figsize=(10,5))
plt.subplot(221)
plt.subplots_adjust(hspace=0.3, wspace=0.5)
plt.grid(linestyle='dotted')
ax = plt.gca()
plt.xlabel('t')
plt.ylabel('Allocation $\\alpha$')
plt.plot([t0,t1],[0,0],linewidth=1,color='grey',linestyle='--')
plt.plot([t0,t1],[1,1],linewidth=1,color='grey',linestyle='--')
plt.plot([t0,t1],[uopt,uopt],linewidth=1, linestyle='dotted', color='black',label='$\\alpha^*_{opt}$')
plt.plot(t[1:],u,linewidth=1,label='$\\alpha$')
ax.set_xlim(t0,t1)
ax2 = ax.twinx()
ax2.set_ylim(ax.get_ylim())
plt.tick_params(axis='y', which='minor', labelleft='off', labelright='on')
plt.yticks([uopt], ['$\\alpha^*_{opt}$'])
plt.subplot(222)
plt.grid(linestyle='dotted')
ax = plt.gca()
plt.plot([t0,t1],[popt,popt],linewidth=1, linestyle='dotted', color='black',label='$\hat p^*_{opt}$')
plt.plot(t, p, linewidth=1, label='$p$')
plt.legend()
ax.set_xlim(t0,t1)
plt.xlabel('t')
plt.ylabel('$p$')
ax2 = ax.twinx()
ax2.set_ylim(ax.get_ylim())
plt.tick_params(axis='y', which='minor', labelleft='off', labelright='on')
plt.yticks([popt], ['$p^*_{opt}$'])
plt.subplot(223)
plt.grid(linestyle='dotted')
plt.xlabel('t')
plt.ylabel('Mass fractions')
ax = plt.gca()
ax2 = ax.twinx()
ax2.set_ylim(ax.get_ylim())
plt.tick_params(axis='y', which='minor', labelleft='off', labelright='on')
plt.yticks([rmin,ropt,rmax_old,rmax], ['$r_{min}$','$r^*_{opt}$','$r_{max}$','$r^w_{max}$'])
ax.set_xlim(t[0],t[-1])
plt.fill_between(t, r, color='#374e55', label='$r$', alpha=0.7)
plt.fill_between(t, r, r+m, color='#b24746', label='$m$', alpha=0.8)
plt.fill_between(t, r+m, rmax_old, color='#df8f44', label='$w$', alpha=0.9)
plt.fill_between(t, rmax_old, 1, color='#79af97', label='$q$', alpha=0.9)
plt.plot(t, r, color='#374e55',linewidth=0.5)
plt.plot(t, r+m, color='#b24746',linewidth=0.5)
plt.grid(linestyle='dotted',color='black')
plt.legend(ncol = 2)
plt.subplot(224)
plt.grid(linestyle='dotted')
plt.xlabel('t')
plt.ylabel('Growth rate $\mu$')
ax = plt.gca()
plt.plot([t0,t1],[muopt,muopt],linewidth=1, linestyle='dotted', color='black',label='$\mu^*_{opt}$')
plt.plot(t,mu,linewidth=1,label='$\mu(p,r)$')
ax.set_xlim(t0,t1)
ax2 = ax.twinx()
ax2.set_ylim(ax.get_ylim())
plt.tick_params(axis='y', which='minor', labelleft='off', labelright='on')
plt.yticks([muopt], ['$\mu^*_{opt}$'])
plt.show()
Loading solution: ./problem3.sol